UNEC Journal of Engineering and Applied Sciences Volume 4, No 2, pages 99-105 (2024) Cite this article, 1313 https://doi.org/10.61640/ujeas.2024.1210
Serratia marcescens is a facultative anaerobic gram-negative bacilli. It is motile and belongs to the intestinal microbiota family. It is spread in nature, and can be isolated from various sources including humans, animals, water, soil, insects, and vertebrate gastrointestinal tract [1]. S. marcescens is a saprophytic bacterium capable of withstanding many different environmental conditions with a thermal range from (10-40 °C) and pH between (5-9) [2]. S. marcescens also produces extracellular enzymes such as DNase, gelatinase and lipase [3]. It has the ability to produce catalase but does not have the effectiveness for producing oxidase [4-6].
Some strains of S. marcescens produce the pigment prodigiosin, which causes the red or pink appearance of colonies on nutrient agar [7, 8].
The pathogenicity of S. marcescens depends on the extracellular secreted enzymes, including proteases, fimbria, lipopolysaccharide (LPS) and ShlA hemolysin. These enzymes are considered virulence factors [2, 9]. For example, hemolysins are produced by various pathogenic bacteria and have been proposed to be responsible for their pathogenesis [10, 11]. One type of hemolysin, cytolysin is a group of pore-forming toxins. Cytolysin is typically formed as a homo-oligomer that is integrated into the cell membrane, thereby changing the cell permeability and leading to cell death. Hemolysin is another type of virulence factors, these enzymes have phospholipase C activity [12]. Phospholipise PhlA the other virulence factors is responsible for the lecithinase activity of S. marcescens [{ref:13]. PhlA is cytotoxic when added to epithelial cells and exhibits hemolytic activity, which is due to the accumulation of lysophospholipid cleavage products that can cause membrane instability of target cells [{ref:14},15]. The current study aimed to determine the spread of Enterobacteriaceae virulence genes from S. marcescens strains isolated from pathogenic specimens using conventional PCR.
Sample collection
152 samples were collected from patients suffering from burn infections, wound infections and urinary tract infections during the period from February to June 2023. The samples were collected from Al-Salam Teaching Hospital, Mosul General Hospital, Al-Jumhuriya Educational Hospital, and Mosul Specialized Center for Burns and Cosmetic Treatment in Mosul city.
Isolation and identification
S. marcescens isolates were identified depending on its morphological characteristics on MacConkey, nutrient and DNase agar, in addition to other specific biochemical tests [16]. Identification was further confirmed by VITEK-2 system (bioMérieux, France) as well as 16S rRNA gene sequencing [17]. The primers 27F: AGAGTTTGATCMTGGCTCAG and 1522R: AAGGAGGTGATCCARCCGA were used for the amplification of the 1495bp DNA fragment of the 16S rRNA region [18]. Amplification of 16S rRNA was performed in a DNA thermal cycler, with the following cycling program; Initial denaturation for 3 minutes at 95 oC; followed by 30 cycles including denaturation for 45 seconds at 95 oC, annealing for 30 seconds at 55 oC, extension for 1.5 minute at 72 oC; and a final extension step for 3 minutes at 72oC [19].
DNA extraction:
Genomic DNA was extracted using the DNA Extraction Kit provided by (Geneaid), steps for extractions was done according to the manufacturer's instructions.
Molecular detection of some virulence genes
Primers for eight different virulence genes (table 1) specific for Enterobacteriaceae and S. marcescens were used to search for corresponding genes in S. marcescens using conventional PCR. These genes were: shlA, shlB, phlA, swr, sepA, cnf1, sat and hly. Table 1 shows the size of the virulence genes and the sequence of the primers used in PCR reactions.
Only 5 out of the 152 total samples isolated were identified as S. marcescens according to all identification steps used. The distribution of S. marcescens according to sample types were as follows: UTI 3/68 (4.41%), wound infection 1/46 (2.17%), burn infection 1/38 (2.63). According to our findings, the isolation and diagnostic rate of S. marcescens among other bacterial species was 3.28%. The findings further corroborate the prevalence and existence of this bacterium in all sources of isolation, with a variation in the proportion of isolates from each source.

Figure 1. Distribution of S. marcescens among the rest of the bacterial species in the isolated infections
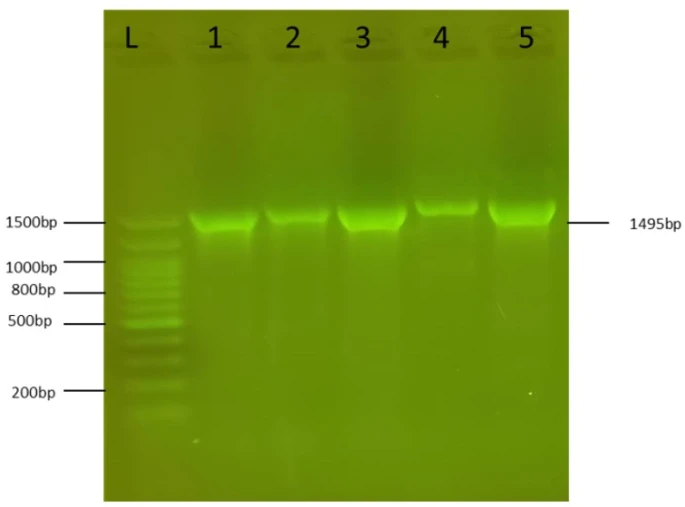
Figure 2. PCR amplification of the 1495bp 16S rRNA fragment from S. marcescens isolates. L:100bp DNA ladder, 1-5: S. marcescens isolate 1-5
The biochemical tests and VITEK 2 system was used to confirm identification of S. marcescens. On MacConkey agar S. marcescens colonies were small, pale and non-lactose fermenters [7]. Colonies were circular convex with a smooth surface on nutrient agar [6]. The biochemical characteristics of S. marcescens were negative for oxidase and positive for catalase. DNase agar (without indicator) showed clear zones around colonies within 5 minutes after HCL addition.
Identification of isolates was confirmed by sequencing their 16S rRNA genes. The PCR results showed that the primers amplified the universal 1495bp band that existed in all five S. marcescens isolates (figure 2). The amplicons were extracted from the agarose gel using the PCR clean-up system (Promega/USA), then sent for DNA sequencing at Psomagen sequencing company (USA). The sequences obtained were BLAST alignment in NCBI and showed high identity to S. marcescens strains. The isolates were submitted in Gene Bank and given the accession numbers OR272225- OR272227.
Analyzing five S. marcescens isolates for the presence of virulence genes revealed the presence of phIA in 80% and hly in 60% of the isolates. The remaining six virulence genes (shlA, shlB, swr, sepA, cnf1, and sat) were not detected. Figure 3 shows the gel electrophoresis picture of hly in three isolates with a size of 123 bp.
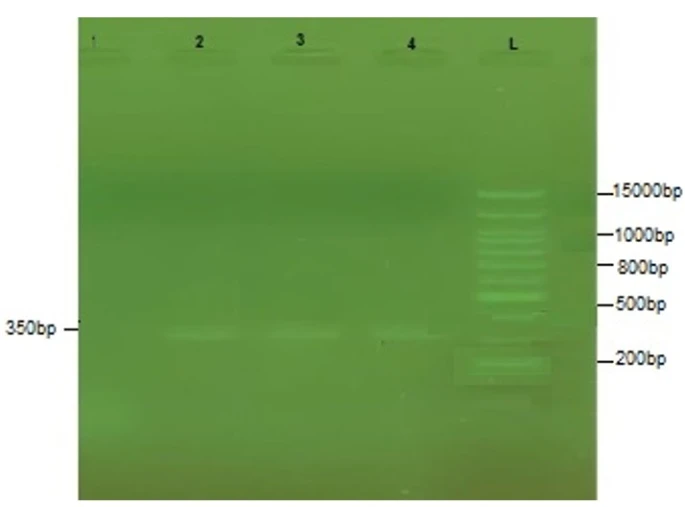
Figure 3. PCR amplification of the hly gene from S. marcescens isolates. L:100bp DNA ladder, 1-4: S. marcescens isolate 1-4
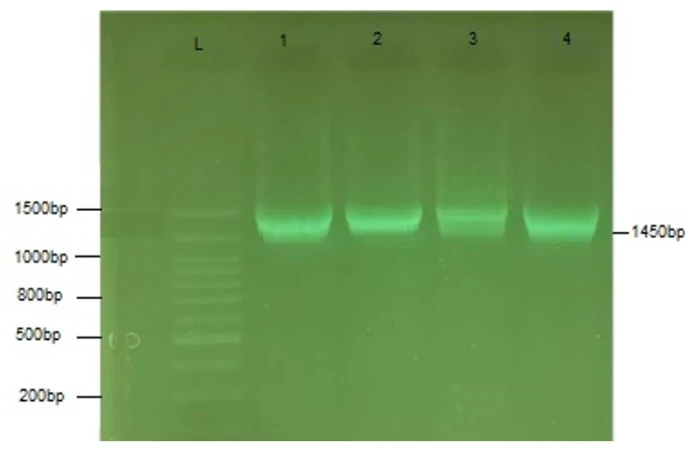
Figure 4. PCR amplification of the 1450bp phIA gene from S. marcescens isolates. L:100bp DNA ladder, 1-4: S. marcescens isolate 1-4
The gel electrophoreses of PhIA for four isolates of S. marcescens are shown in Figure (4). The PhIA size is 1000 bps.
S. marcescens is a common opportunistic pathogen in hospitalized patients. Serratia spp. is usually non-pigmented and causes pneumonia, bacteremia, and endocarditis specifically in narcotics addicts. The isolation of S. marscecens is most common in UTI, followed by burns and wounds. Our findings are supported by earlier research [19]. Although S. marcescens is recognized to cause UTI, mainly in health care settings and in patients with urinary catheters, the bacterial variables implicated in UTI remain unknown for this organism [20].
The diagnostic findings for the five bacterial isolates were received and firmly verified that they were S. marcescens, particularly because they had 98% similarity to the S. marcescens registered internationally inside NCBI. The 16S rRNA gene was used to confirm identification of S. marcescens isolates. 16S rRNA has a truncation length of 1495 bps. Observations reveal that the nucleotide sequence is not mutable and only slowly varied over time [21].
PhIA was found in four of the isolates under investigation, and this finding was consistent with earlier researches [22, 23]. Given the numerous well-characterized instances of secreted phospholipases with involvement in bacterial virulence, the possibility of PhlA contributing to S. marcescens pathogenicity is an appealing concept [24]. However, the hly has been found in three different bacterial clones.
The rest of the genes did not appear during molecular diagnosis. The loss of virulence factor genes in bacteria is a multifaceted phenomenon documented in the scientific literature. Spontaneous mutations, genetic recombination, and selective pressures play pivotal roles in this process [25, 26]. Environmental influences, host immunological reactions, and competition with other microbes all impose selective pressures on bacterial genomes, resulting to adaptive alterations [27]. The loss of plasmids and transposon excision are examples of horizontal gene transfer processes that further deplete virulence components in bacterial populations [23, 28]. Furthermore, the loss of virulence factors has been related to chromosomal rearrangements, including deletions and rearrangements [29]. Bacterial virulence factors evolve as a result of dynamic interactions fostered by long-term host-pathogen coevolution, thereby changes in their sequence might prevent the primers from binding to their specific sites [30]. Future studies are required to sequence genomes from local Serratia isolates and study the variations in virulence genes if any and identify if such changes prevent primer annealing.
In Iraq, the number of infections with S. marcescens bacterium is steadily growing. Identifying virulence factor genes is critical for detecting mutations. Two virulence factor genes out of eight were found among the isolated strains of S. marcescens. The discovery of novel strains of these bacteria alerts, cautions, and draws the attention of the health system.
1 Z. Kahrarian, S. Khazayel, A. Tajehmiri, Asian Journal of Research in Medicine and Medical Science 1(1) (2019) 18.
2 A. Hejazi, and F.R. Falkiner, Journal of medical microbiology 46(11) (1997) 903. https://doi.org/10.1099/00222615 -46-11-903
3 R. Zivkovic Zaric, M. Zaric, M. Sekulic, N. Zornic, J. Nesic, V. Rosic, T. Vulovic, M. Spasic, M. Vuleta, J. Jovanovic, D. Jovanovic, S. Jakovljevic, P. Canovic, Antibiotics 12(2) (2023) 367. https://doi.org/10.3390/antibiotics12020367
4 M.H. Abd-Alla, S.R. Bashandy, S. Schnell, S. Ratering, Phytoparasitica 39(2) (2011) 175. https://doi.org/10.1007/s12600 -011-0148-6
5 E.W. Koneman, W.C. Winn, S.D. Allen, W.M. Janda, G.W. Procop, P.C. Schreckenberger, G.L. Woods, Color Atlas & Textbook of Diagnostic Microbiology.6th Edition, Lippincot Williams &Wilkins, Philadelphia , USA (2006).
6 F. Grimont, and P.A.D. Grimont, The prokaryotes 6 (2006) 197. https://doi.org/10.1007/0-387-30746-X_9
7 R.L. Ferreira, G.S. Rezende, M.S.F. Damas, M. Oliveira-Silva, A. Pitondo-Silva, M.C.A. Brito, E. Leonardecz, F.R. de Góes, E.B. Campanini, I. Malavazi, A.F. da Cunha, M.D.S. Pranchevicius, Frontiers in microbiology 11 (2020) 956. https://doi.org/10.3389/fmicb.2020.00956
8 D. Kljakić, M.Z. Milosavljević, M. Jovanović, V.Č. Popović, S. Raičević, Open medicine (Wars) 16(1) (2020) 81. https://doi.org/10.1515/med-2021-0205
9 R. Hertle, Current protein & peptide science 6(4) (2005) 313. https://doi.org/10.2174/1389203054546370
10 M. Palmer, Toxicon: official journal of the International Society on Toxinology 39(11) (2001) 1681. https://doi.org/10.1016/s0041 -0101(01)00155-6
11 D.H. Walker, H.M. Feng, V.L. Popov, The American journal of tropical medicine and hygiene 65(6) (2001) 936. https://doi.org/10.4269/ajtmh.2001.65.936
12 J. Sakurai, M. Nagahama, M. Oda, Journal of biochemistry 136(5) (2004) 569. https://doi.org/10.1093/jb/mvh161
13 M. Givskov, L. Olsen, S. Molin, Journal of bacteriology 170(12) (1988) 5855. https://doi.org/10.1128/jb.170.12.5855 -5862.1988
14 J.K. Song, M.K. Kim, J.S. Rhee, Journal of biotechnology 72(1-2) (1999) 103. https://doi.org/10.1016/s0168 -1656(99)00096-6
15 M. Labbate, H. Zhu, L. Thung, R. Bandara, M.R. Larsen, M.D. Willcox, M. Givskov, S. A. Rice, S. Kjelleberg, Journal of bacteriology 189(7) (2007) 2702. https://doi.org/10.1128/JB.01582 -06
16 G.M. Garrity, J. Bell, G. Lilburn, Taxonomic Outline of the Prokaryotes. In: Bergey’s manual of Systemic Bacteriology, 2nd ed., Berlin Heidelberg, New York (2005).
17 A.M. Khaleel, R.M. Faisal, H.A. Altaii, Revista Bionatura 8(3) (2023) 113. http://dx.doi.org/10.21931/RB/2023.08.03.113
18 A.M. Khaleel, R.M. Faisal, H.A. Altaii, Malaysian Journal of Microbiology 19(2) (2023) 115. http://dx.doi.org/10.21161/mjm.220105
19 R. Abdulrazzaq, R. Faisal, Journal of Life and Bio Sciences Research 3(01) (2022) 01. http://dx.doi.org/10.38094/jibsr30151
20 L.H. Su, J.T. Ou, H.S. Leu, P.C. Chiang, Y.P. Chiu, J.H. Chia, A.J. Kuo, C.H. Chiu, C. Chu, T.L. Wu, C.F. Sun, T.V. Riley, B.J. Chang, Infection Control Group, Journal of clinical microbiology 41(10) (2003) 4726. https://doi.org/10.1128/JCM.41.10.4726 - 4732.2003
21 J.E. Clarridge 3rd, Clinical microbiology reviews 17(4) (2004) 840. https://doi.org/10.1128/CMR.17.4.840 -862.2004
22 K. Shimuta, M. Ohnishi, S. Iyoda, N. Gotoh, N. Koizumi, H. Watanabe, BMC microbiology 9 (2009) 1. https://doi.org/10.1186/1471 -2180-9-261
23 J. Hacker, E. Carniel, EMBO reports 21(51) (2001) 376. https://doi.org/10.1093/embo - reports/kve097
24 M.T. Anderson, L.A. Mitchell, H.L.T. Mobley, Journal of bacteriology 199(16) (2017) e00159-17. https://doi.org/10.1128/JB.00159 -17
25 M. Flores-Díaz, L. Monturiol-Gross, C. Naylor, A. Alape-Girón, A. Flieger, Microbiology and molecular biology reviews: MMBR 80(3) (2016) 597. https://doi.org/10.1128/MMBR.00082 -15
26 E. Dadachova, A. Casadevall, Current opinion in microbiology 11(6) (2008) 525. https://doi.org/10.1016/j.mib.2008.09.013
27 J. Wiedenbeck, F.M. Cohan, FEMS microbiology reviews 35(5) (2011) 957. https://doi.org/10.1111/j.1574 -6976.2011.00292.x
28 M. Diard, E. Bakkeren, J.K. Cornuault, K. Moor, A. Hausmann, M.E. Sellin, C. Loverdo, A. Aertsen, M. Ackermann, M. De Paepe, E. Slack, W.D. Hardt, Science (New York, N.Y.) 355(6330) (2017) 1211. https://doi.org/10.1126/science.aaf8451
29 C.M. Johnson, A.D. Grossman, Annual review of genetics 49 (2015) 577. https://doi.org/10.1146/annurev -genet-112414-055018
30 H. Ochman, J. Lawrence, E. Groisman, Nature 405 (2000) 299. https://doi.org/10.1038/35012500
31 R. Medzhitov, D.S. Schneider, M.P. Soares, Science (New York, N.Y.) 335(6071) (2012) 936. https://doi.org/10.1126/science.1214935
M.I. Khalil, M.A. Al-Tobje, R.M. Faisal, Molecular detection of virulence genes of Serratia marcescens isolates from diverse clinical sources, UNEC J. Eng. Appl. Sci. 4(2) (2024) 99-105 https://doi.org/10.61640/ujeas.2024.1210
Anyone you share the following link with will be able to read this content:

This article is licensed under the Creative Commons Attribution ( CC BY 4.0 ) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Z. Kahrarian, S. Khazayel, A. Tajehmiri, Asian Journal of Research in Medicine and Medical Science 1(1) (2019) 18.
A. Hejazi, and F.R. Falkiner, Journal of medical microbiology 46(11) (1997) 903. https://doi.org/10.1099/00222615 -46-11-903
R. Zivkovic Zaric, M. Zaric, M. Sekulic, N. Zornic, J. Nesic, V. Rosic, T. Vulovic, M. Spasic, M. Vuleta, J. Jovanovic, D. Jovanovic, S. Jakovljevic, P. Canovic, Antibiotics 12(2) (2023) 367. https://doi.org/10.3390/antibiotics12020367
M.H. Abd-Alla, S.R. Bashandy, S. Schnell, S. Ratering, Phytoparasitica 39(2) (2011) 175. https://doi.org/10.1007/s12600 -011-0148-6
E.W. Koneman, W.C. Winn, S.D. Allen, W.M. Janda, G.W. Procop, P.C. Schreckenberger, G.L. Woods, Color Atlas & Textbook of Diagnostic Microbiology.6th Edition, Lippincot Williams &Wilkins, Philadelphia , USA (2006).
F. Grimont, and P.A.D. Grimont, The prokaryotes 6 (2006) 197. https://doi.org/10.1007/0-387-30746-X_9
R.L. Ferreira, G.S. Rezende, M.S.F. Damas, M. Oliveira-Silva, A. Pitondo-Silva, M.C.A. Brito, E. Leonardecz, F.R. de Góes, E.B. Campanini, I. Malavazi, A.F. da Cunha, M.D.S. Pranchevicius, Frontiers in microbiology 11 (2020) 956. https://doi.org/10.3389/fmicb.2020.00956
D. Kljakić, M.Z. Milosavljević, M. Jovanović, V.Č. Popović, S. Raičević, Open medicine (Wars) 16(1) (2020) 81. https://doi.org/10.1515/med-2021-0205
R. Hertle, Current protein & peptide science 6(4) (2005) 313. https://doi.org/10.2174/1389203054546370
M. Palmer, Toxicon: official journal of the International Society on Toxinology 39(11) (2001) 1681. https://doi.org/10.1016/s0041 -0101(01)00155-6
D.H. Walker, H.M. Feng, V.L. Popov, The American journal of tropical medicine and hygiene 65(6) (2001) 936. https://doi.org/10.4269/ajtmh.2001.65.936
J. Sakurai, M. Nagahama, M. Oda, Journal of biochemistry 136(5) (2004) 569. https://doi.org/10.1093/jb/mvh161
M. Givskov, L. Olsen, S. Molin, Journal of bacteriology 170(12) (1988) 5855. https://doi.org/10.1128/jb.170.12.5855 -5862.1988
J.K. Song, M.K. Kim, J.S. Rhee, Journal of biotechnology 72(1-2) (1999) 103. https://doi.org/10.1016/s0168 -1656(99)00096-6
M. Labbate, H. Zhu, L. Thung, R. Bandara, M.R. Larsen, M.D. Willcox, M. Givskov, S. A. Rice, S. Kjelleberg, Journal of bacteriology 189(7) (2007) 2702. https://doi.org/10.1128/JB.01582 -06
G.M. Garrity, J. Bell, G. Lilburn, Taxonomic Outline of the Prokaryotes. In: Bergey’s manual of Systemic Bacteriology, 2nd ed., Berlin Heidelberg, New York (2005).
A.M. Khaleel, R.M. Faisal, H.A. Altaii, Revista Bionatura 8(3) (2023) 113. http://dx.doi.org/10.21931/RB/2023.08.03.113
A.M. Khaleel, R.M. Faisal, H.A. Altaii, Malaysian Journal of Microbiology 19(2) (2023) 115. http://dx.doi.org/10.21161/mjm.220105
R. Abdulrazzaq, R. Faisal, Journal of Life and Bio Sciences Research 3(01) (2022) 01. http://dx.doi.org/10.38094/jibsr30151
L.H. Su, J.T. Ou, H.S. Leu, P.C. Chiang, Y.P. Chiu, J.H. Chia, A.J. Kuo, C.H. Chiu, C. Chu, T.L. Wu, C.F. Sun, T.V. Riley, B.J. Chang, Infection Control Group, Journal of clinical microbiology 41(10) (2003) 4726. https://doi.org/10.1128/JCM.41.10.4726 - 4732.2003
J.E. Clarridge 3rd, Clinical microbiology reviews 17(4) (2004) 840. https://doi.org/10.1128/CMR.17.4.840 -862.2004
K. Shimuta, M. Ohnishi, S. Iyoda, N. Gotoh, N. Koizumi, H. Watanabe, BMC microbiology 9 (2009) 1. https://doi.org/10.1186/1471 -2180-9-261
J. Hacker, E. Carniel, EMBO reports 21(51) (2001) 376. https://doi.org/10.1093/embo - reports/kve097
M.T. Anderson, L.A. Mitchell, H.L.T. Mobley, Journal of bacteriology 199(16) (2017) e00159-17. https://doi.org/10.1128/JB.00159 -17
M. Flores-Díaz, L. Monturiol-Gross, C. Naylor, A. Alape-Girón, A. Flieger, Microbiology and molecular biology reviews: MMBR 80(3) (2016) 597. https://doi.org/10.1128/MMBR.00082 -15
E. Dadachova, A. Casadevall, Current opinion in microbiology 11(6) (2008) 525. https://doi.org/10.1016/j.mib.2008.09.013
J. Wiedenbeck, F.M. Cohan, FEMS microbiology reviews 35(5) (2011) 957. https://doi.org/10.1111/j.1574 -6976.2011.00292.x
M. Diard, E. Bakkeren, J.K. Cornuault, K. Moor, A. Hausmann, M.E. Sellin, C. Loverdo, A. Aertsen, M. Ackermann, M. De Paepe, E. Slack, W.D. Hardt, Science (New York, N.Y.) 355(6330) (2017) 1211. https://doi.org/10.1126/science.aaf8451
C.M. Johnson, A.D. Grossman, Annual review of genetics 49 (2015) 577. https://doi.org/10.1146/annurev -genet-112414-055018
H. Ochman, J. Lawrence, E. Groisman, Nature 405 (2000) 299. https://doi.org/10.1038/35012500
R. Medzhitov, D.S. Schneider, M.P. Soares, Science (New York, N.Y.) 335(6071) (2012) 936. https://doi.org/10.1126/science.1214935