UNEC Journal of Engineering and Applied Sciences Volume 3, No 2, pages 21-32 (2023) Cite this article, 1499 https://doi.org/10.61640/ujeas.2023.1203
Polycyclic Aromatic Hydrocarbons (PAHs) organic compounds composed of carbon and hydrogen. They are ubiquitous, harmful to the environment and to people's health by being a strong carcinogenic agent to the lungs, cervix, bladder, breast, and prostate, as well as causing bone loss, eye irritation, and skin sensitivity [1]. PAHs are derived from three sources; biogenic, pyrogenic and petrogenic origins. While living organisms produce the biogenic PAHs, the pyrogenic sources are derived from the thermal decomposition of organic matter (fat, carbohydrate, protein), including those made during processing foods at high temperatures [2,3]. Incomplete combustion from natural (forest and brush fires) and man-made combustion sources (automobile emissions and cigarette smoke) produce the PAHs. Higher hydrophobicity and resistance to microbial degradation are associated with the increased number of fused rings [4]. Due to their high molecular stability, PAHs tend to remain and circulate between environmental compartments for extended periods of time. However, they present low or no solubility in water, which, alongside their resistance to biodegradation, makes them susceptible to adsorption onto suspended particles and/or sedimentation, creating a direct exposure route to benthic biota. Due to their lipophilicity and their ability to interact with lipid cell membranes, they usually show bio-accumulative characteristics [5,6]. PAHs in the environment can be degraded through various processes such as photolysis, volatilization, chemical degradation and bioremediation. Both metabolic reactions, aerobic oxidation and anaerobic dechlorination, are known to be involved in the degradation of PCBs by microorganisms [7]. The degradation of biphenyl usually begins with the breakdown to form catechol. The catechol will be further degraded to form acetyl-CoA and succinyl-CoA, which subsequently be incorporated into the citric acid cycle where the organism can obtain energy from its oxidation. The series of degradation processes finally degrades the hydrocarbon to produce carbon dioxide and water, which are the safest end products [4]. The biphenyl operons, known as bph genes, are comprised of genes which encode the enzymes required for biphenyl metabolism that are usually separated on different operons [8]. This study aimed to isolate biphenyl degraders from different types of soils in Iraq and indicate their ability to metabolize biphenyl by growth curves and molecular detection of aromatic degrading genes.
Collection of samples
Fifty five soil samples were collected from reeds soil, sandy soil, landfill soil, oil refinery soil, agricultural soil, sulfur station soil, garden soil, and sewage contaminated soil. Soil samples were collected from different locations in Mosul - Iraq at a depth of 1-10 cm during the period from September, 2021 to February, 2022. The samples were delivered to the laboratory in sterile plastic containers and kept at room temperature to be used for further experiments.
Isolation of biphenyl degrading bacteria
One gram of soil sample was inoculated in a 250ml flask containing 50ml DSMZ minimal medium [9]. Flasks were inoculated in duplicates and incubated at 28°C and 37°C for 7-10 days. Flasks were daily examined for turbidity. Serial dilutions were prepared for flasks that showed turbidity on DSMZ plates containing biphenyl added as crystals on the lid of the plate. Colonies were sub-cultured on DSMZ plates supplemented with biphenyl for the purpose of purification and verification. Growing colonies were streaked on DSMZ plates lacking biphenyl as a control step to avoid autotrophic bacteria [10]. Bacterial strains were preserved in nutrient broth containing 20% glycerol and stored at -80°C for long term storage.
Molecular identification
Genomic DNA was isolated from biphenyl degraders using Geneaid Genomic DNA purification kit (Taiwan) following the recommended steps by Geneaid company. Genomic DNA was used as a template to amplify the 16S rRNA gene using the primers 27F (5’ AGAGTTTGATCMTGGCTCAG 3’) and 1522R (5’ AAGGAGGTGATCCARCCGCA 3’) used by [11] and following the conditions used elsewhere [12]. The degenerate primers BPHD-f3 (AACTGGAARTTYGCIGCVGA) and BPHD-r1 (ACCCAGTTYTCICCRTCGTC) was used to amplify a 524bp DNA fragment from the Rieske non-heme iron dioxygenase gene following the conditions used in table 1 [13]. The second set of degenerate primers, DOalpha, was designed to target a 264bp fragment from angular dioxygenase genes by using the primers DOalpha-2 (5’ TGYHSNTAYCAYGGNTGG 3’) and DOalpha-3 (5’ TCNRCNGCNARYTTCCARTT 3’) using the conditions shown in table 2 [14]. PCR was conducted using Promega master mix in 20µl reactions and the DNA ladder supplied from New England Biolabs (USA) was used to identify the size of the amplified fragments. Amplified DNA was purified directly from PCR products or from agarose gel using PCR clean-up kit (Promega-USA). Purified DNA samples were sent for sequencing at Psomagene sequencing company (Maryland-USA). Sequences were blasted against the DNA sequence references submitted to NCBI database to detect the identity of the isolate [15].
Growth curve experiment
The growth rate for biphenyl degrading isolates was determined using growth curve experiments. Biphenyl degraders were grown in 250ml flasks containing 50ml DSMZ minimal media and incubated in a shaker incubator (110 rpm) at 37°C. DSMZ medium was supplemented with 3mM biphenyl which was added as crystals. Growth curve experiment was conducted in triplicates and optical densities were measured at 600nm. Graphpad Prism (v.8.0.2) was used to draw growth curves and all required statistics were calculated automatically [16].
Isolation of biphenyl degrading bacteria
DSMZ medium incubated at 28°C did not show growth of any biphenyl degrading bacteria. However, a soil sample that was incubated on 37°C showed signs of turbidity after 7 days of incubation. Serial dilutions were spread from this culture on DSMZ agar plates. The bacterial strains isolated was grown on nutrient agar plates and their cultural characteristics was recorded. These strains were also stained with Gram stain to identify their type of pigmentation as seen in Table 4.
Both Gram positive and Gram negative biphenyl degraders were detected. Gram positive isolates belonged to Rhodococcus and Microbacterium. Two species were detected from the genus Rhodococcus, R. pyridinovorans and R. rhodochrous. These strains were mostly colorful (orange to red or pink), except Rhodococcus sp. RM5 which was the only member from Rhodococcus that had a white creamy appearance. Creamy appearance of Rhodococcus was detected earlier by [17]. However, multiple researchers worldwide have shown the colorful properties of Rhodococcus on mineral media. The pthalate degrading Rhodococcus sp. 2G was shown to produce orange to pink colonies when grown on LB agar [18]. This bacterium was also shown to degrade biphenyl due to acquisition of all required biphenyl degrading genes that were detected by analyzing its genome [19].
A single species from the Gram positive Microbacterium, M. barkeri, was isolated for its ability to degrade biphenyl, this bacterium had a light yellow color of colonies. Microbacterium has been previously isolated for its ability to degrade biphenyl, this bacterium was isolated from landfills and was shown to carry biphenyl degrading capabilities [20].
Three genera from Gram negative bacteria, Achromobacter, Extensimonas, and Pseudomonas was isolated for their ability to metabolize biphenyl. Cultural characteristics showed that Achromobacter had light yellow colonies when grown on nutrient agar plates. These results agree with [21] whom isolated this bacteria from soil contaminated with oil. Pseudomonas luteola was isolated from sewage samples for their ability to metabolize biphenyl. The colonies had an orange to yellow color when grown on DSMZ containing biphenyl. Similar cultural characteristics was observed for this bacterium isolated from a sewage sample from an electricity generator company located in Baghdad-Iraq [22]. Extensimonas perlucida was the third Gram negative bacterium that was isolated for its ability to use biphenyl as a sole source for carbon and energy. This bacterium showed circular transparent colonies when grown on DSMZ agar containing biphenyl (figure 4-9). Similar cultural characteristics was observed by [23] whom isolated this bacterium for the first time from a sludge sample of a pesticide treating plant.
Molecular identification using 16S universal primers
The nine cultures that showed turbidity in DSMZ liquid media containing biphenyl and produced colonies on DSMZ plates with biphenyl and not on control plates were shown to contain a pure culture of biphenyl degraders. Genomic DNA was isolated from these cultures and their 16S rRNA gene was amplified. As expected a product of 1495bp was detected on agarose gel as shown in figure (1).
The PCR products were purified and sequenced, then the sequence was used as a query in BLAST searches at the website of the National center for Biotechnology Information (https://blast.ncbi.nlm.nih.gov/). Results showed that these biphenyl degraders belonged to five different bacterial genera; Rhodococcus, Achromobacter, Microbacterium, Extensimonas, and Pseudomonas as shown in table 5. All biphenyl degraders were submitted to NCBI and given a corresponding accession number. Each strain when submitted was given a strain name composed of the initial RM except RM4 that we did not submit due to its bad sequencing results. This strain was identified according to its cultural characteristics.
Molecular detection of biphenyl degrading genes
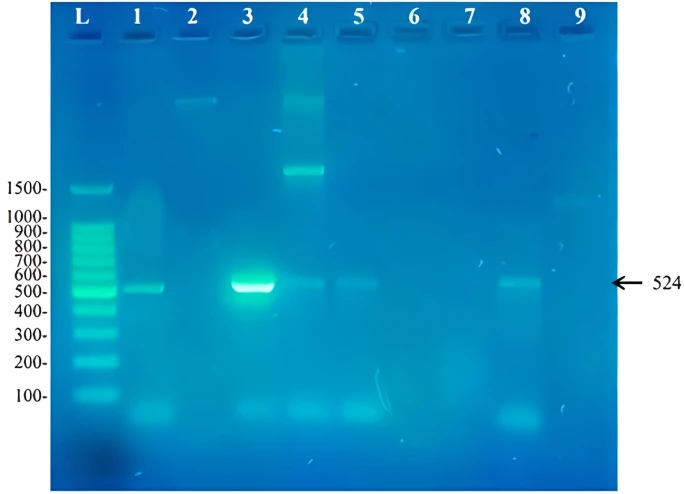
Two sets of primers were used to identify the presence of biphenyl degrading genes in the isolates understudy. These primers were degenerate primers that were designed by researchers working in this field for the purpose of identifying aromatic degrading genes in isolates that are not yet sequenced. The primer BPHD-f3 and BPHD-r1 was used to amplify a 524bp DNA fragment from the Rieske non-heme iron dioxygenase gene expected to be used in the initial step of biphenyl degradation. Our results showed that the presence of this gene in these biphenyl degrading strains (figure 2). The isolates that contained this gene was Rhodococcus pyridinivorans RM2, Extensimonas perlucida RM1, Rhodococcus sp. RM4, Rhodococcus sp. RM5 and Rhodococcus pyridinivorans RM8. The presence of the gene in these bacterial genera show that these primers amplify Rieske non-heme iron dioxygenase gene from both Gram negative and Gram positive bacteria. When using this set of primers by [13], they showed that six out of the seven novel genes sequenced have not yet been structurally studied yet and if there sequences where known before their work, that would have helped in designing degenerate primers that that have better quality. This work shows that the bacteria capable of degrading biphenyl that did not show a corresponding band may obtain a novel biphenyl dioxygenase sequence that needs to be discovered in different ways. No research has been found for Extensimonas perlucida as this study was the first study worldwide that shows the ability of this bacterium to metabolize biphenyl as a sole source for carbon and energy. Extensimonas perlucida is a Gram negative bacterium that belongs to the family Comamonadaceae [24]. Along with the species Extensimonas perlucida, the genus Extensimonas contains another member known as E. vulgaris [23]. E. vulgaris was first isolated in China from a sewage sample contaminated with industrial waste and was shown to resist 48°C [25]. According to the fact that industrial waste is usually composed of oil residues as an contaminant, it is expected from Extensimonas members to carry PAH degrading capabilities. Growth of Extensimonas perlucida at relatively high temperatures (48°C) explains how Extensimonas perlucida is capable of metabolizing biphenyl at 37°C [24].
Figure 2. Amplicons of a 524bp fragment from the Rieske non-heme iron dioxygenase found in biphenyl degrading strains from this study. L: 100bp DNA ladder, 1: Rhodococcus pyridinivorans RM2, 2: Rhodococcus rhodochrous RM3, 3: Extensimonas perlucida RM1, 4
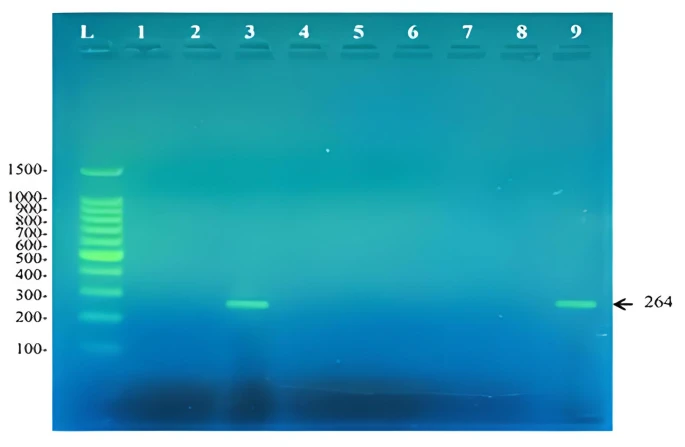
The second set of primers (DOalpha-2 and DOalpha-3) was designed to target angular dioxygenase genes involved in the first step of aromatic hydrocarbon degradation other than biphenyl. Angular dioxygenation is the attack of a certain aromatic compound at the C-C bond where one of the carbons is involved in the bridge between the two benzene rings [26]. This type of dioxgenation attack is involved in the oxygenation of dibenzo-p-dioxin [27], carbazole [28], our results showed that E. perlucida and Pseudomonas luteola RM9 contains an angular dioxygenase gene that is probably involved in degrading other aromatic compounds besides biphenyl. This was noticed by the ability of the DOalpha set of primers, designed to target genes encoding angular dioxygenase enzymes, to amplify a 264bp fragment as shown in figure 3.
Figure 3. Amplicons of a 264bp fragment from the angular dioxygenase amplified using the primers DOalpha -2 and DOalpha-3. L: 100bp DNA ladder, 1: Rhodococcus pyridinivorans RM2, 2: Rhodococcus rhodochrous RM3, 3: Extensimonas perlucida RM1, 4: Rhodococcu
Biphenyl degrading ability of isolates understudy
The growth rate of the nine biphenyl degraders understudy was identified by growing the strains individually in DSMZ broth medium and measuring the absorbance OD600 at different time points. Growth rates varied with different strains reflecting the efficiency of their enzymes in degrading biphenyl. The growth rate (doubling time) was 24.79 hours for E. perlucida RM1, 17.05 hours for Rh. pyridinivorans RM2, 17.69 hours for Rh. rhodochrous RM3, 16.80 hours for Rhodococcus sp. RM4, 19.43 for Rhodococcus sp. RM5, 19.65 for Achromobacter sp. RM6, 16.48 hours for M. barkeri RM7, 16.93 for Rh. pyridinovorans RM8 and 16.99 for P. luteola RM9 (figures 4-12, respectively). Duplication of the strains on DSMZ containing biphenyl as a sole source for carbon and energy provides another proof for the ability of the strains understudy to metabolize biphenyl.
Figure 4. Growth of E. perlucida RM1 on 3mM biphenyl in DSMZ medium. Each point reflects a triplicate and the error bars represents the standard deviation of the mean
Figure 5. Growth of Rh. pyridinivorans RM2 on 3mM biphenyl in DSMZ medium. Each point reflects a triplicate and the error bars represents the standard deviation of the mean
Figure 6. Growth of Rh. rhodochrous RM3 on 3mM biphenyl in DSMZ medium. Each point reflects a triplicate and the error bars represents the standard deviation of the mean
Figure 7. Growth of Rhodococcus sp. RM4 on 3mM biphenyl in DSMZ medium. Each point reflects a triplicate and the error bars represents the standard deviation of the mean
Figure 8. Growth of Rhodococcus sp. RM5 on 3mM biphenyl in DSMZ medium. Each point reflects a triplicate and the error bars represents the standard deviation of the mean
Figure 9. Growth of Achromobacter sp. RM6 on 3mM biphenyl in DSMZ medium. Each point reflects a triplicate and the error bars represents the standard deviation of the mean
Figure 10. Growth of M. barkeri RM7 on 3mM biphenyl in DSMZ medium. Each point reflects a triplicate and the error bars represents the standard deviation of the mean
Figure 11. Growth of Rh. pyridinovorans RM8 on 3mM biphenyl in DSMZ medium. Each point reflects a triplicate and the error bars represents the standard deviation of the mean
Figure 12. Growth of P. luteola RM9 on 3mM biphenyl in DSMZ medium. Each point reflects a triplicate and the error bars represents the standard deviation of the mean
Polycyclic Aromatic Hydrocarbons are ubiquitous environmental pollutants that are harmful to the environment and to community health. Biphenyl is a member of this group that has several detrimental effects especially when chlorinated. The search for new biphenyl degraders helps in providing more efficient bio-degraders that can eliminate this toxic aromatic compound when found in the environment thereby providing better bioremediation results. This work successfully isolated several Gram-negative and Gram-positive biphenyl degraders from Iraqi soil in addition to introducing new biphenyl degraders to the field that were not previously known worldwide. The ability of these biphenyl degraders were confirmed by their acquisition of multiple biphenyl degrading genes involved in the biphenyl degradation pathway.
1 G. Goudarzi, N. Alavi, A.A. Babaei, S. Geravandi, E. Idani, S. Salmanzadeh, M. J. Mohammadi, Polycyclic Aromatic Compounds 42(5) (2022) 1978.
2 I.W. Ofosu, E.A. Larbi, D. Alale, G.M. Ankar-Brewoo, H.E. Lutterodt, Journal of Food and Nutrition Research 10(7) (2022) 467.
3 R.M. Faisal, R.S. Alsaffar,. Bioplastic Degradation, Rafidain Journal of Science 32(2) (2023) 69.
4 S. Adenan, Ch.F. Wong, S.S.A.A. Azziz, S.C.S. Nang, R. Misnan, D.U. Lamasudin, B.Y.Ch. Lau, Malaysian Journal of Microscopy 18(1) (2022) 66.
5 J.V. de Pinho, P.d.A. Rodrigues, I.D.L. Guimarães, F.C. Monteiro, R.G. Ferrari, R.A. Hauser-Davis, & C.A. Conte-Junior, International journal of environmental research and public health 19(3) (2022) 1211.
6 R.M. Faisal, Doc. diss.: Understanding the role of Dibenzofuran 4, 4a dioxygenase reveals a silent pathway for biphenyl degradation in Sphingomonas wittichii RW1 and helps in engineering dioxin degrading strains. Rutgers University-School of Graduate Studies (2019).
7 T. Hara, & Y. Takatsuka, Canadian Journal of Microbiology 68(3) (2022) 191.
8 F. Khalid, M.Z. Hashmi, N. Jamil, A. Qadir, & M.I. Ali, Environmental Science Pollution Research 28(9) (2021) 10474. https://doi.org/10.1007/s11356-020-11996-2
9 E.K. Camboim, A.P. Almeida, M.Z. Tadra-Sfeir, F.G. Junior, P.P. Andrade, C. S. McSweeney, & F. Riet-Correa, The Scientific World Journal 2012 (2012). https://doi.org/10.1100/2012/178254
10 A.J. Roberts, Doc. diss.: Genetic and functional characterization of biphenyl, diphenylmethane, and diphenylether degradation. Rutgers University (2020).
11 A.M. Khaleel, R.M. Faisal, & H.A. Altaii, Malaysian Journal of Microbiology 19(2) (2023a) 115. http://dx.doi.org/10.21161/mjm.220105
12 R. Abdulrazzaq, & R. Faisal, Journal of Life and Bio Sciences Research 3(01) (2022) 01. http://dx.doi.org/10.38094/jibsr30151
13 S. Iwai, B. Chai, W.J. Sul, J.R. Cole, S.A. Hashsham, & J.M. Tiedje, The ISME journal 4(2) (2010) 279.
14 T. Iida, Y. Mukouzaka, K. Nakamura, & T. Kudo, Applied and Environmental Microbiology 68(8) (2022) 3716.
15 A.M. Khaleel, R.M. Faisal, H.A. Altaii, Revis Bionatura 8(3) (2023b) 113. http://dx.doi.org/10.21931/RB/2023.08.03.113
16 R.M. Faisal, & A.H. Rasol, Journal of Genetic Engineering and Biotechnology 20(1) (2022) 1.
17 N. Yadav, & A. Yadav, Biodivers Int J 3(2) (2019) 37
18 J. Hou, F. Liu, N. Wu, J. Ju, & B. Yu, Journal of nanobiotechnology 14(1) (2016) 1.
19 H.-M. Zhao, R.-W. Hu, H. Du, X.-P. Xin, Y.-W. Li, H. Li, Q.-Y. Cai, C.-H. Mo, J.-S. Liu, & D.-M. Zhou, Science of the Total Environment 640 (2018) 646.
20 H.-M. Zhao, R.-W. Hu, H.-B. Huang, H.-F. Wen, H. Du, Y.-W. Li, H. Li, Q.-Y. Cai, C.-H. Mo, & J.-S. Liu, Biology and Fertility of Soils 53(6) (2017) 663.
21 E. Kaczorek, K. Sałek, U. Guzik, B. Dudzińska-Bajorek, & A.Olszanowski, International Biodeterioration & Biodegradation 78 (2013) 7.
22 F.K. Emran, H.A. Walli, F.J. Shinjar, & M.A. Abdulameer, In Journal of Physics: Conference Series, Iraq 1294(7) (2019) 072014.
23 Q. Peng, M. Sheng, Z. Yang, H. Ni, Q. Li, Y. Li, & J. He, Current Microbiology 77(7) (2020) 1316.
24 P.E. Rouvière, & M.W. Chen, FEMS microbiology letters 227(1) (2003) 101.
25 W.Y. Zhang, M.X. Fang, W.W. Zhang, C. Xiao, X.Q. Zhang, Z.P. Yu, & M. Wu, International journal of systematic and evolutionary microbiology 63(Pt_6) (2013) 2062.
26 P.V. Bünz, & A.M. Cook, Journal of bacteriology 175(20) (1993) 6467.
27 S. Saibu, S.A. Adebusoye, & G. O.Oyetibo, Bioresources and Bioprocessing 7(1) (2020) 1.
28 L.B. Salam, M.O. Ilori, O.O. Amund, M. Numata, T. Horisaki, & H. Nojiri, Environmental Science and Pollution Research 21(15) (2014) 9311.
R.M. Sobhi, R.M. Faisal, Distribution of biphenyl degraders in soil samples in Mosul-Iraq, UNEC J. Eng. Appl. Sci. 3(2) (2023) 21-32 https://doi.org/10.61640/ujeas.2023.1203
Anyone you share the following link with will be able to read this content:

This article is licensed under the Creative Commons Attribution ( CC BY 4.0 ) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
G. Goudarzi, N. Alavi, A.A. Babaei, S. Geravandi, E. Idani, S. Salmanzadeh, M. J. Mohammadi, Polycyclic Aromatic Compounds 42(5) (2022) 1978.
I.W. Ofosu, E.A. Larbi, D. Alale, G.M. Ankar-Brewoo, H.E. Lutterodt, Journal of Food and Nutrition Research 10(7) (2022) 467.
R.M. Faisal, R.S. Alsaffar,. Bioplastic Degradation, Rafidain Journal of Science 32(2) (2023) 69.
S. Adenan, Ch.F. Wong, S.S.A.A. Azziz, S.C.S. Nang, R. Misnan, D.U. Lamasudin, B.Y.Ch. Lau, Malaysian Journal of Microscopy 18(1) (2022) 66.
J.V. de Pinho, P.d.A. Rodrigues, I.D.L. Guimarães, F.C. Monteiro, R.G. Ferrari, R.A. Hauser-Davis, & C.A. Conte-Junior, International journal of environmental research and public health 19(3) (2022) 1211.
R.M. Faisal, Doc. diss.: Understanding the role of Dibenzofuran 4, 4a dioxygenase reveals a silent pathway for biphenyl degradation in Sphingomonas wittichii RW1 and helps in engineering dioxin degrading strains. Rutgers University-School of Graduate Studies (2019).
T. Hara, & Y. Takatsuka, Canadian Journal of Microbiology 68(3) (2022) 191.
F. Khalid, M.Z. Hashmi, N. Jamil, A. Qadir, & M.I. Ali, Environmental Science Pollution Research 28(9) (2021) 10474. https://doi.org/10.1007/s11356-020-11996-2
E.K. Camboim, A.P. Almeida, M.Z. Tadra-Sfeir, F.G. Junior, P.P. Andrade, C. S. McSweeney, & F. Riet-Correa, The Scientific World Journal 2012 (2012). https://doi.org/10.1100/2012/178254
A.J. Roberts, Doc. diss.: Genetic and functional characterization of biphenyl, diphenylmethane, and diphenylether degradation. Rutgers University (2020).
A.M. Khaleel, R.M. Faisal, & H.A. Altaii, Malaysian Journal of Microbiology 19(2) (2023a) 115. http://dx.doi.org/10.21161/mjm.220105
R. Abdulrazzaq, & R. Faisal, Journal of Life and Bio Sciences Research 3(01) (2022) 01. http://dx.doi.org/10.38094/jibsr30151
S. Iwai, B. Chai, W.J. Sul, J.R. Cole, S.A. Hashsham, & J.M. Tiedje, The ISME journal 4(2) (2010) 279.
T. Iida, Y. Mukouzaka, K. Nakamura, & T. Kudo, Applied and Environmental Microbiology 68(8) (2022) 3716.
A.M. Khaleel, R.M. Faisal, H.A. Altaii, Revis Bionatura 8(3) (2023b) 113. http://dx.doi.org/10.21931/RB/2023.08.03.113
R.M. Faisal, & A.H. Rasol, Journal of Genetic Engineering and Biotechnology 20(1) (2022) 1.
N. Yadav, & A. Yadav, Biodivers Int J 3(2) (2019) 37
J. Hou, F. Liu, N. Wu, J. Ju, & B. Yu, Journal of nanobiotechnology 14(1) (2016) 1.
H.-M. Zhao, R.-W. Hu, H. Du, X.-P. Xin, Y.-W. Li, H. Li, Q.-Y. Cai, C.-H. Mo, J.-S. Liu, & D.-M. Zhou, Science of the Total Environment 640 (2018) 646.
H.-M. Zhao, R.-W. Hu, H.-B. Huang, H.-F. Wen, H. Du, Y.-W. Li, H. Li, Q.-Y. Cai, C.-H. Mo, & J.-S. Liu, Biology and Fertility of Soils 53(6) (2017) 663.
E. Kaczorek, K. Sałek, U. Guzik, B. Dudzińska-Bajorek, & A.Olszanowski, International Biodeterioration & Biodegradation 78 (2013) 7.
F.K. Emran, H.A. Walli, F.J. Shinjar, & M.A. Abdulameer, In Journal of Physics: Conference Series, Iraq 1294(7) (2019) 072014.
Q. Peng, M. Sheng, Z. Yang, H. Ni, Q. Li, Y. Li, & J. He, Current Microbiology 77(7) (2020) 1316.
P.E. Rouvière, & M.W. Chen, FEMS microbiology letters 227(1) (2003) 101.
W.Y. Zhang, M.X. Fang, W.W. Zhang, C. Xiao, X.Q. Zhang, Z.P. Yu, & M. Wu, International journal of systematic and evolutionary microbiology 63(Pt_6) (2013) 2062.
P.V. Bünz, & A.M. Cook, Journal of bacteriology 175(20) (1993) 6467.
S. Saibu, S.A. Adebusoye, & G. O.Oyetibo, Bioresources and Bioprocessing 7(1) (2020) 1.
L.B. Salam, M.O. Ilori, O.O. Amund, M. Numata, T. Horisaki, & H. Nojiri, Environmental Science and Pollution Research 21(15) (2014) 9311.